Research
In Silico Drug Discovery
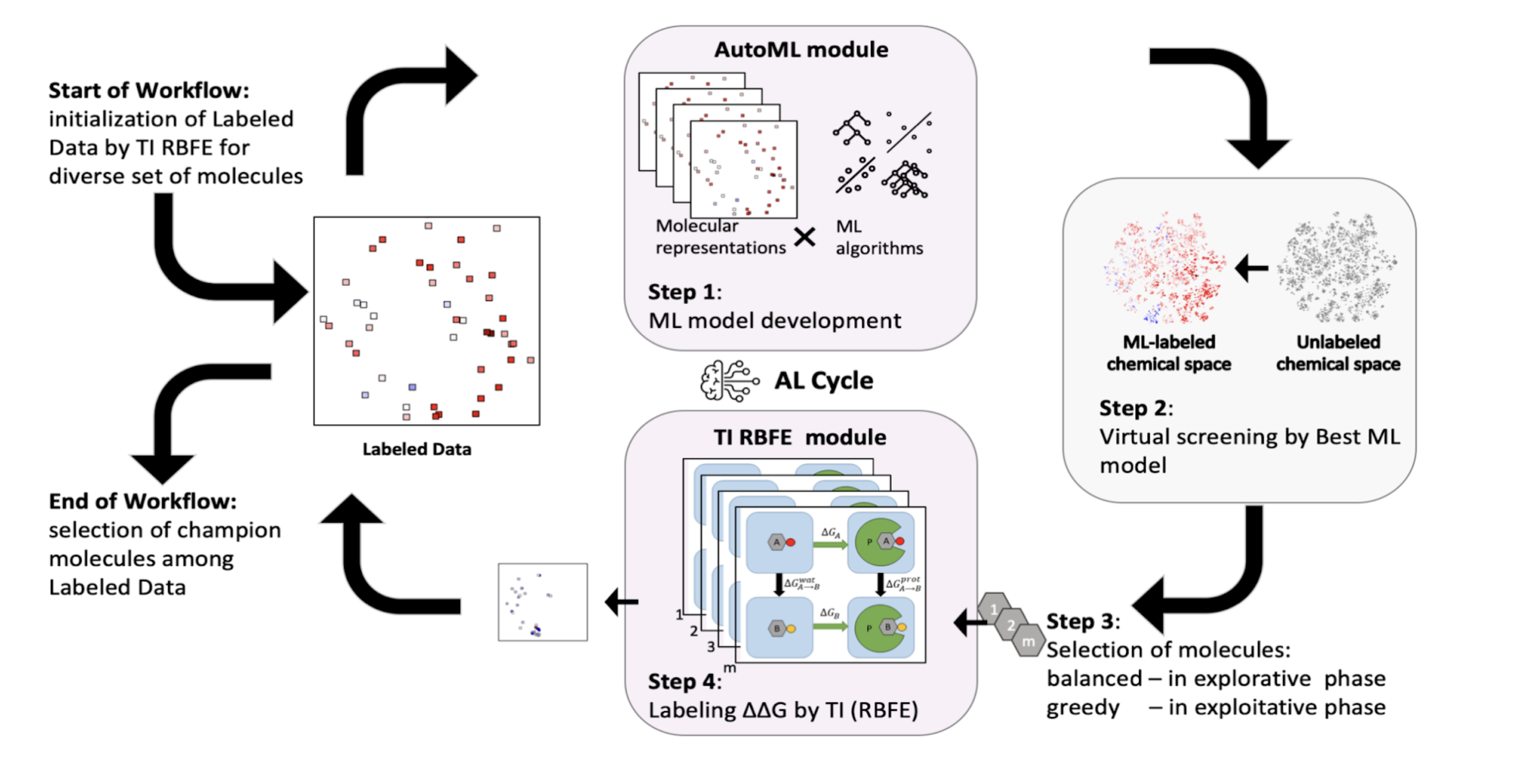
We have developed several high-throughput binding free energy simulation workflows for drug discovery. We utilize high-throughput absolute binding free energy simulation for hit discovery and relative binding free energy simulations combined with our active learning workflow for lead optimization. Combined, we utilized these workflows to win 1st place in the CACHE Challenge #1, an international computational drug discovery competition, where we and our collaborators discovered 14 novel inhibitors to the challenging leucine-rich repeat kinase 2 (LRRK2) WDR domain, an understudied Parkinson's disease drug target with no known molecular inhibitors prior to our work.
- Active Learning Guided Drug Design Lead Optimization Based on Relative Binding Free Energy Modeling., Gusev F, Gutkin E, Kurnikova MG, Isayev O., J. Chem. Inf. Model. 63(2) pp 583–594, (2023) ↗
- In silico screening of LRRK2 WDR domain inhibitors using deep docking and free energy simulations, Gutkin, E; Gusev, F; Gentile, F; Ban, F; Koby, SB; Narangoda, C; Isayev, O; Cherkasov, A; Kurnikova, M.G., DOI: 10.1039/D3SC06880C, Chem. Sci., Advance Article (2024) ↗
Gating Mechanism and Ion Channel Block in Kainate Receptors
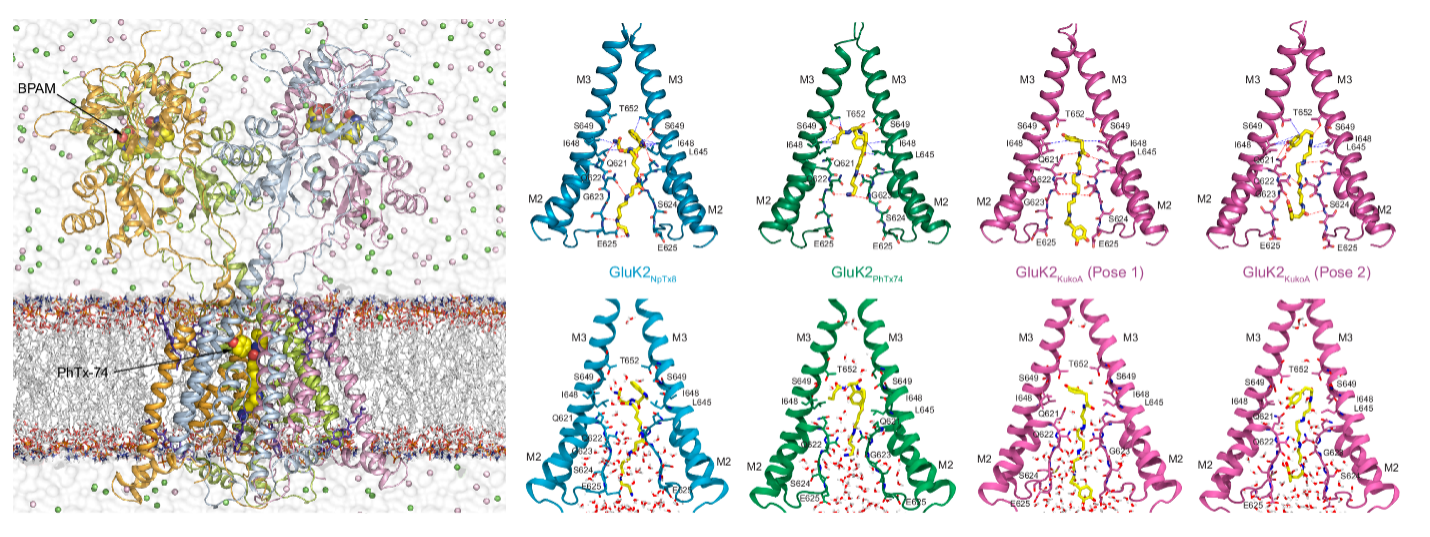
Kainate receptors (KARs) are another subtype of iGluR channels, and despite their importance in excitatory neurotransmission, the mechanism of KAR gating remains poorly understood due to the lack of available structures. In addition, calcium-permeable KARs undergo ion channel block, but the therapeutic potential of channel blockers is also underdeveloped. Our research on GluK2 KARs investigates the channel opening and gating mechanism of these receptors with computational methods. We use extensive molecular dynamics simulations to explore the conformational flexibility of the receptor and the effects of specific ion channel blockers on gating and ion conduction. Since GluK2 KARs are linked to several neurological conditions, including epilepsy, schizophrenia, and autism, understanding their dynamic behavior has a therapeutic importance. Our work seeks to provide atomic-level insight into KAR function and its differences from closely related iGluR family members, expanding the broader knowledge of glutamate receptor biophysics.
- Gangwar SP, Yelshanskaya MV, Aktolun M, Yen LY, Newton TP, Strømgaard K, Kurnikova MG, Sobolevsky AI. Trapping of spermine, Kukoamine A, and polyamine toxin blockers in GluK2 kainate receptor channels. Nat Commun 15, 10257 (2024). ↗
- Gangwar SP, Yelshanskaya MV, Nadezhdin KD, Yen LY, Newton TP, Aktolun M, Kurnikova MG, Sobolevsky AI. Kainate receptor channel opening and gating mechanism. Nature 630, 762-768 (2024) . ↗
Integrating Molecular Dynamics and Machine Learning to Unravel AMPA Receptor Conductance Mechanisms
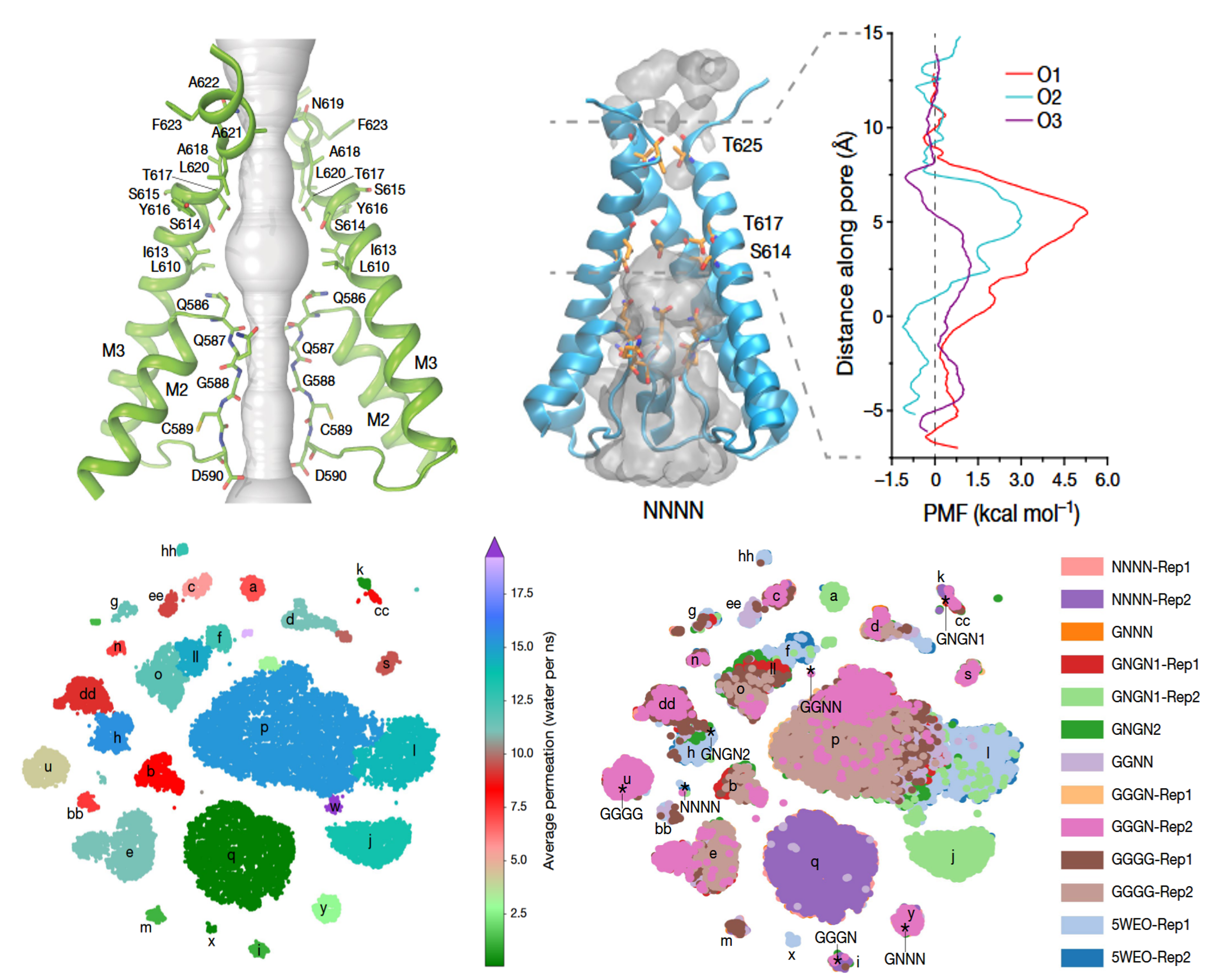
This project focuses on understanding how AMPA receptor channels transition between distinct subconductance states and how these states influence water permeability through the pore. To address this, we combine large-scale molecular dynamics simulations with data-driven analyses, including clustering and machine learning, to uncover the structural features that govern ion and water flow at different conductance levels. These features include local helicity, inter-residue distances, pore geometry, and hydration patterns, which serve as descriptors of the receptor’s conformational landscape. By integrating such structural, dynamical, and functional descriptors into predictive models, we aim to identify the parameters most strongly correlated with permeation properties and to reveal the collective mechanisms underlying subconductance transitions. This combination of physics-based simulation with machine learning provides a quantitative framework for linking conformational ensembles to functional outcomes, offering new insights into the molecular determinants of channel gating and conductance modulation.
- Yelshanskaya MV, Patel DS, Kottke CM, Kurnikova MG, Sobolevsky AI. Opening of glutamate receptor channel to subconductance levels. Nature 605, 172–178 (2022) ↗
- M. V. Yelshanskaya, S. Mesbahi-Vasey, M. G. Kurnikova and A. I. Sobolevsky, “Role of the ion channel extracellular collar in AMPA receptor gating”, Scientific Reports 7: 1050 (2017). ↗
Catalytic mechanism of α-ketoglutarate (AKG)-dependent enzymes

In collaboration with Prof. Guo’s lab, we are investigating the detailed molecular mechanisms of α-ketoglutarate (AKG)-dependent non-heme iron enzymes. This large and versatile family of proteins catalyzes a wide range of chemical reactions. Our long-term objective is to enable the rational design of AKG-dependent enzymes capable of efficiently driving desired chemical transformations.
At present, our efforts center on HrmJ protein variants, which exhibit shifts in chemical specificity—switching between hydroxylation, cyclization, and halogenation—depending on specific amino acid mutations. To rationalize these changes in enzymatic reactivity, we are performing extensive molecular dynamics simulations to probe the stability and dynamics of different catalytic states.
To further characterize the active enzyme states, we are developing next-generation machine-learned interatomic potentials (MLIPs) tailored for the iron-coordinated active centers of AKG-dependent proteins. These MLIPs deliver quantum-level accuracy at a fraction of the computational cost, providing a powerful framework for understanding and ultimately engineering this important class of enzymes.
- Shan Xue, Yijie Tang, Igor V. Kurnikov, Hsuan-Jen Liao, Jikun Li, Nei-Li Chan, Maria G. Kurnikova, Wei-chen Chang, Yisong Guo, Chapter Nine - Spectroscopic and computational studies of a bifunctional iron- and 2-oxoglutarate dependent enzyme, AsqJ, ↗
NMDA receptor transmembrane domain structure and function
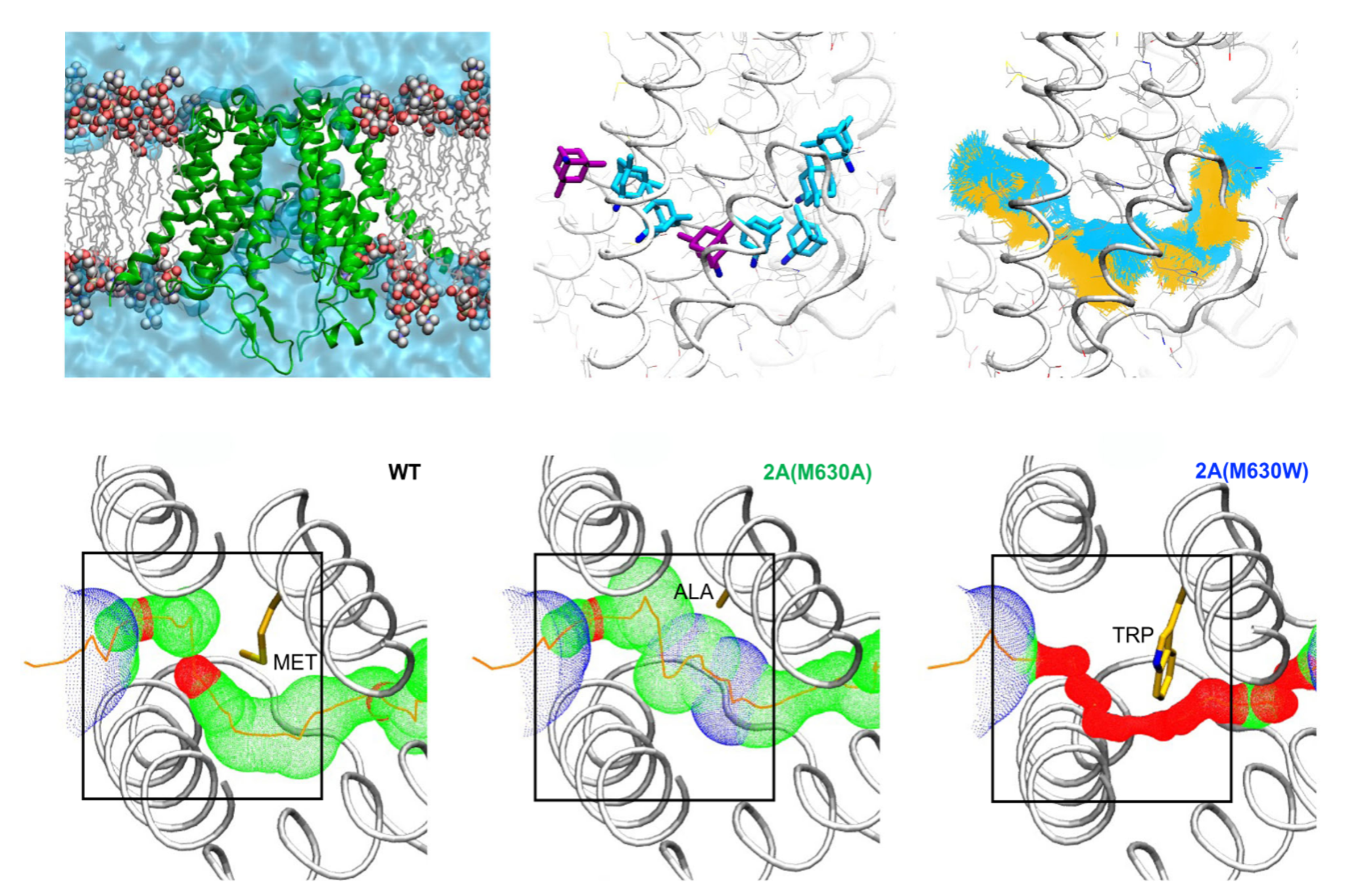
N-methyl-D-aspartate receptors (NMDARs) are glutamate- and glycine-gated ion channels that are essential for synaptic plasticity, learning, and memory. Dysregulation of NMDAR function has been linked to a range of neurological and psychiatric disorders, making them an important target for mechanistic and therapeutic studies. Our research focuses on the transmembrane domain of NMDARs, where we investigate the molecular basis of membrane-to-channel inhibition (MCI), a mode of open-channel block in which small molecules enter from the lipid bilayer and interact with the pore to inhibit ion conduction, a line of work we pursue in collaboration with Jon W. Johnson’s group.
To address these questions, we use molecular dynamics simulations and free energy calculations to characterize the conformational landscape of the channel and to quantify ligand binding energetics. We are particularly interested in open-channel blockers such as memantine, MK-801 and its structural analogues, which access the pore through defined pathways within the membrane. By mapping these permeation routes and calculating the energetic barriers associated with channel entry and binding, we aim to identify the structural and dynamical features that govern inhibitor potency and selectivity.
This work provides an atomistic description of how small-molecule inhibitors engage the NMDAR pore and modulate conductance. By linking ligand permeation pathways to energetic landscapes of inhibition, we seek to uncover the fundamental principles of open-channel block, offering mechanistic insights that complement experimental electrophysiology and guide the development of new therapeutic strategies targeting glutamatergic signaling.
- Wilcox, M.R., Nigam, A., Glasgow, N.G. et al. Inhibition of NMDA receptors through a membrane-to-channel path. Nat Commun 13, 4114 (2022). ↗
- S. Mesbahi-Vasey, J. W. Johnson, M. G. Kurnikova, “Reaction coordinate for an ion association with a protein binding site: configurations of the ligand coordination shell”, submitted. ↗
- S. Mesbahi-Vasey, L. Veras, M. Yonkunas, J. W. Johnson, M. G. Kurnikova, “All atom NMDA receptor transmembrane domain model development and simulations in lipid bilayers and water”, PLoS ONE 12(6): e0177686 (2017). ↗
Conformational Dynamics of Gating and Auxiliary Modulation of AMPA Receptors
We focus on the dynamics of the AMPA receptor (AMPAR) gating and modulation by auxiliary subunits. AMPARs mediate fast excitatory neurotransmission in the brain, and their regulation strongly influences synaptic plasticity. They are an important member of ionotropic glutamate receptor (iGluR) family and are implicated in many neurodegenerative diseases which make them key drug targets. Our molecular dynamic simulations are designed to capture conformational changes in key domains, particularly the ligand-binding domain and the transmembrane gating machinery. Additionally, we study the role of auxiliary proteins, including TARPs and CNIH2, in stabilizing functional states and modulating ion permeation. By systematically characterizing these structural mechanisms, we aim to contribute to a better understanding of how AMPARs support learning and memory at the molecular level.
- Yelshanskaya MV, Patel DS, Kottke CM, Kurnikova MG, Sobolevsky AI. Opening of glutamate receptor channel to subconductance levels. Nature 605, 172–178 (2022) ↗
Ion Permeation and Gating Mechanisms of the Transient Receptor Potential (TRP) Channels
Our work on TRP channels focuses on understanding the dynamical and functional features of this ion channel, which plays essential roles in cellular magnesium homeostasis and signaling by controlling the permeation of cations such as Zn2+, Mg2+, and Ca2+ ions. Although they are linked to a wide range of disorders, the molecular mechanism of ion permeation and the selective modulation is still not fully understood. We use molecular dynamics simulations to study how structural dynamics regulate their gating and ion permeation as well as how small molecules or mutations affect their selective modulation. In our recent collaborative work with an inhibitor molecule, anticancer agent CCT128930, we showed that the inhibitor binds into the vanilloid-like site of TRPM7 and stabilizes the channel in a closed, non-conducting state. MD simulations also revealed the key interactions between the receptor residues and the ligand, highlighting the central role of the binding site for the selective interaction of TRPM7 with small molecules, shedding light on future drug design.
- Nadezhdin KD, Correia L, Shalygin A, Aktolun M, Neuberger A, Gudermann T, Kurnikova MG, Chubanov V, Sobolevsky AI. Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930. Cell Rep 43(4), 114108 (2024). ↗
- Nadezhdin KD, Correia L, Narangoda C, Patel DS, Neuberger A, Gudermann T, Kurnikova MG, Chubanov V, Sobolevsky AI. Structural mechanisms of TRPM7 activation and inhibition. Nat Commun 14, 2639 (2023). ↗
- Sakipov S, Sobolevsky AI, Kurnikova MG. Ion Permeation Mechanism in Epithelial Calcium Channel TRVP6. Sci Rep 8, 5715 (2018). ↗