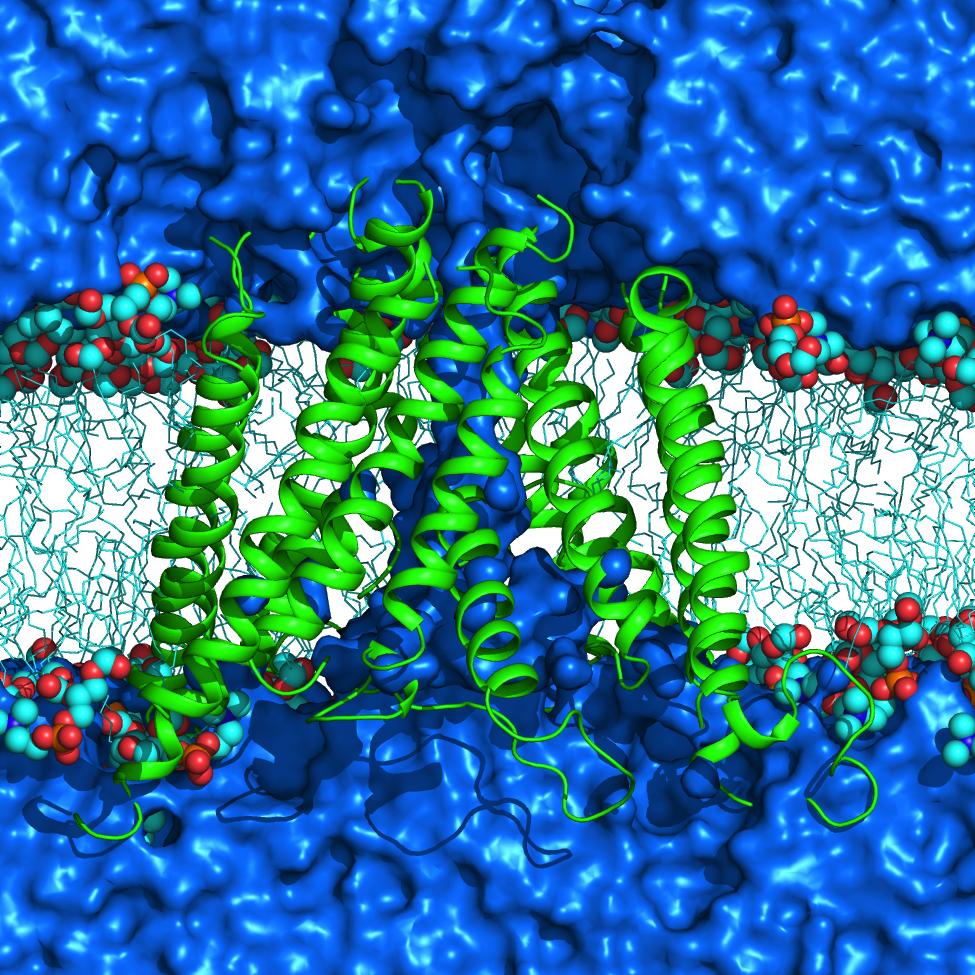
NMDA receptors (NMDARs) are glutamate- and glycine-gated ionotropic glutamate receptors (iGluRs) found mainly in postsynaptic membranes in the central nervous system. NMDARs are required for many forms of synaptic plasticity, including long-term potentiation and long-term depression, physiological processes that are involved in learning and memory. Dysfunction of NMDARs has been associated with multiple nervous system disorders. We investigate a variety of properties in the transmembrane domain (TMD) of NMDARs.
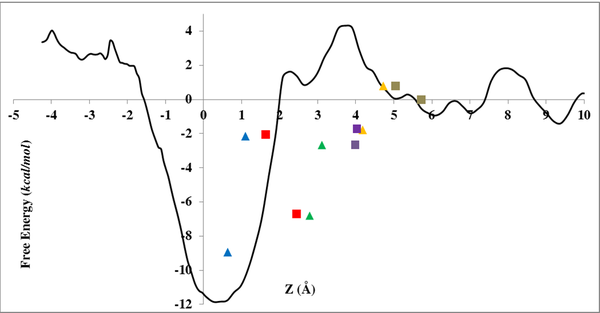
One area of interest includes study of ion selectivity in NMDARs. NMDAR channels are permeable to monovalent cations including Na+ and K+, as well as Ca2+, and are blocked by Mg2+ in a membrane potential-dependent manner. Using molecular dynamics simulations and free energy calculations, we are investigating the basis for Ca2+ permeation versus Mg2+ block in NMDARs. This work led to proposing a novel reaction coordinate (RC) for the calculation of the potential of mean force (PMF) inside narrow channels. This new RC takes into account the slowest degree of freedom for ion permeation reaction, a property lacking when using a straightforward Z-coordinate, a common RC used for ion permeation PMF calculation inside ion channels. Currently, we are exploring the full configurational space of the Mg2+ and Ca2+ in NMDAR channel to accomplish an accurate PMF for describing permeation versus block of divalent ions.
Another area of research, in collaboration with Jon W. Johnson's group, is interaction of several drugs with NMDARs. NMDARs can be inhibited in several ways, one of which is open channel block, in which molecules bind in the pore of NMDARs and block ion flow. Memantine (Mem), a drug used to treat Alzheimer’s disease, is an NMDAR open channel blocker that binds in the NMDAR pore. We study the interaction of Mem with NMDARs and investigate its permeation pathway to the binding site.
Within the M2 region of each NMDAR subunit lies a remarkably conserved tryptophan (W) that can be found at the homologous site in a variety of channels and receptors in phylogenetically distant organisms. In NMDARs, this highly conserved W is located at the interface between the M2 region and the adjacent M3 region. The conserved W plays a role in NMDAR function; mutation of the conserved W in the GluN2B subunit (W607) severely diminishes Mg2+ block. However, mutating the homologous residue in GluN1 subunits (W608) does not affect Mg2+ block, but alters desensitization. To further explore the function of the conserved W in NMDARs, in collaboration with Jon W. Johnson's group, we examined GluN1/GluN2A receptors with mutations at the conserved W and surrounding residues. Our work provided molecular-detailed understanding of altered function of NMDAR seen in electrophysiological experiments by our collaborators.
Recent Publications:
S. Mesbahi-Vasey, J. W. Johnson, M. G. Kurnikova, “Reaction coordinate for an ion association with a protein binding site: configurations of the ligand coordination shell”, submitted.
S. Mesbahi-Vasey, L. Veras, M. Yonkunas, J. W. Johnson, M. G. Kurnikova, “All atom NMDA receptor transmembrane domain model development and simulations in lipid bilayers and water”, PLoS ONE 12(6): e0177686 (2017).
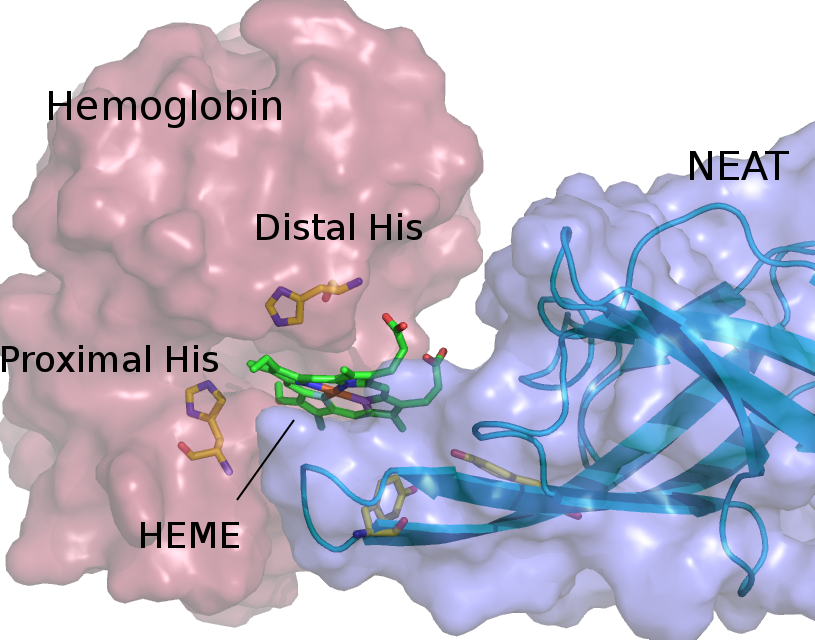
Red blood cell hemolysis in sickle cell disease (SCD) releases free hemoglobin. Extracellular hemoglobin and its degradation products, free heme and iron, are highly toxic due to oxidative stress induction and decrease in nitric oxide availability. We propose an approach that helps to eliminate extracellular hemoglobin toxicity in SCD by employing a bacterial protein system that evolved to extract heme from extracellular hemoglobin. NEAr heme Transporter (NEAT) domains from iron-regulated surface determinant proteins from Staphylococcus aureus specifically bind free heme as well as facilitate its extraction from hemoglobin. We demonstrate that a purified NEAT domain fused with human haptoglobin β-chain is able to remove heme from hemoglobin and reduce heme content and peroxidase activity of hemoglobin. We further use molecular dynamics (MD) simulations to resolve molecular pathway of heme transfer from hemoglobin to NEAT, and to elucidate molecular mechanism of such heme transferring process. Our study is the first of its kind, in which simulations are employed to characterize the process of heme leaving hemoglobin and subsequent rebinding with a NEAT domain. Our MD results highlight important amino acid residues that facilitate heme transfer and will guide further studies for the selection of best NEAT candidate to attenuate free hemoglobin toxicity.
Publications:
Sakipov S, Rafikova O, Kurnikova MG, Rafikov R. Molecular mechanisms of bio-catalysis of heme extraction from hemoglobin Redox Biol. 11:516-523. (2017)
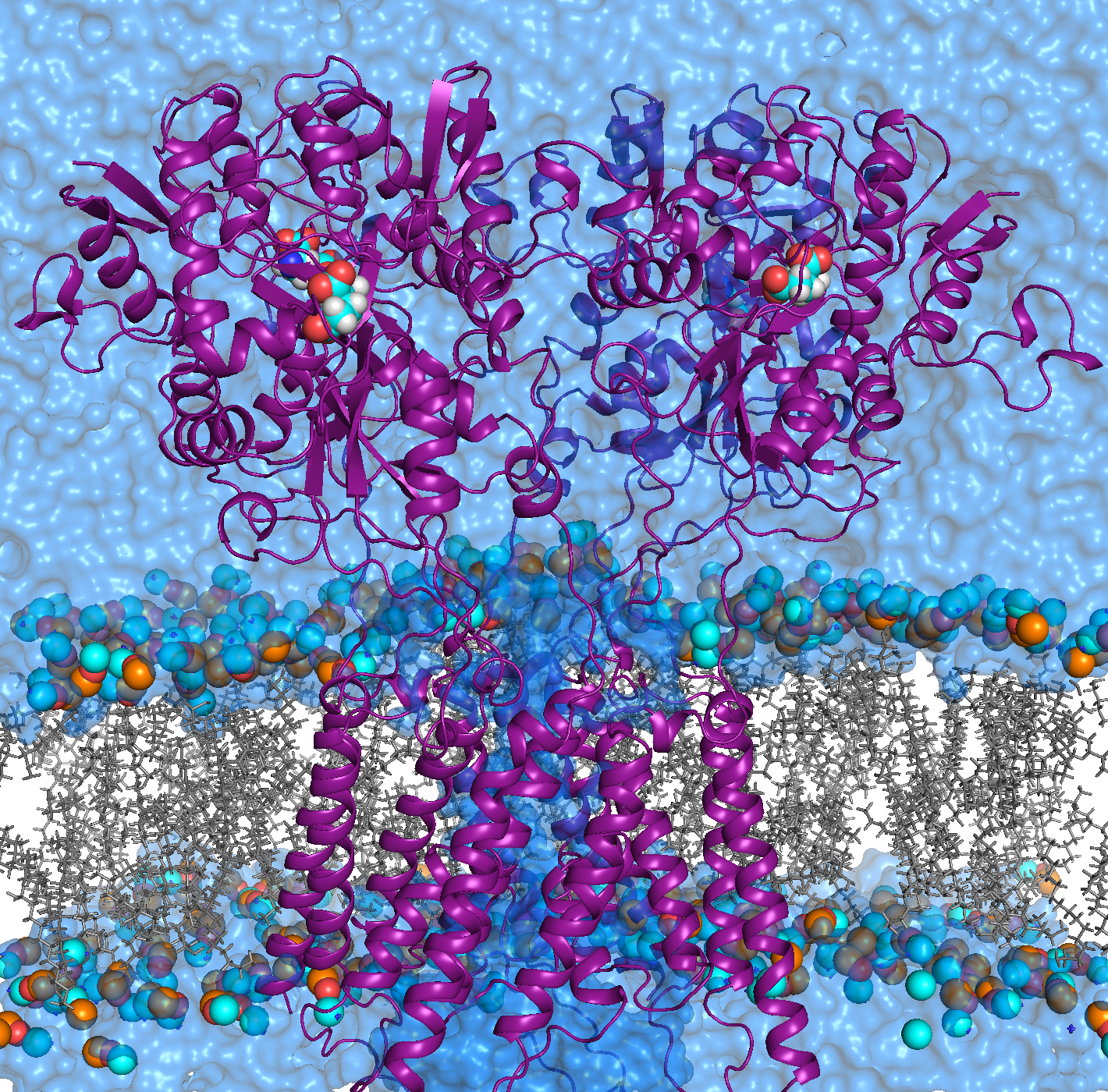
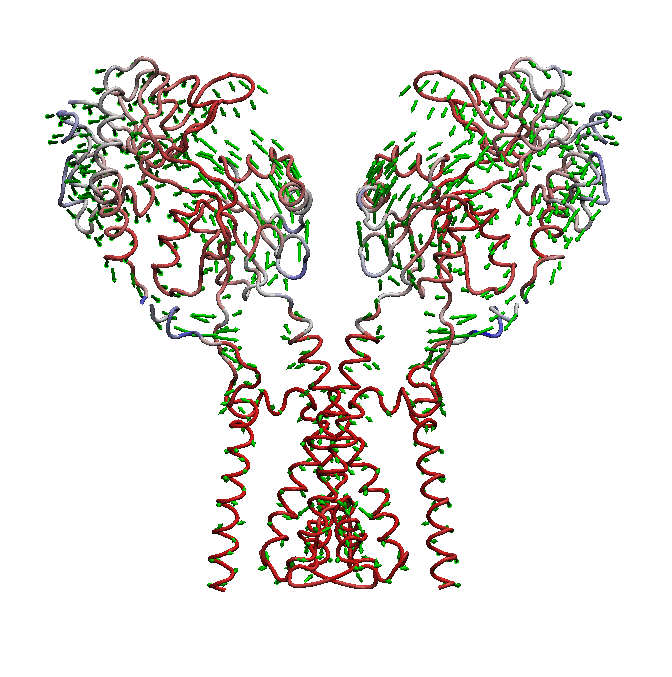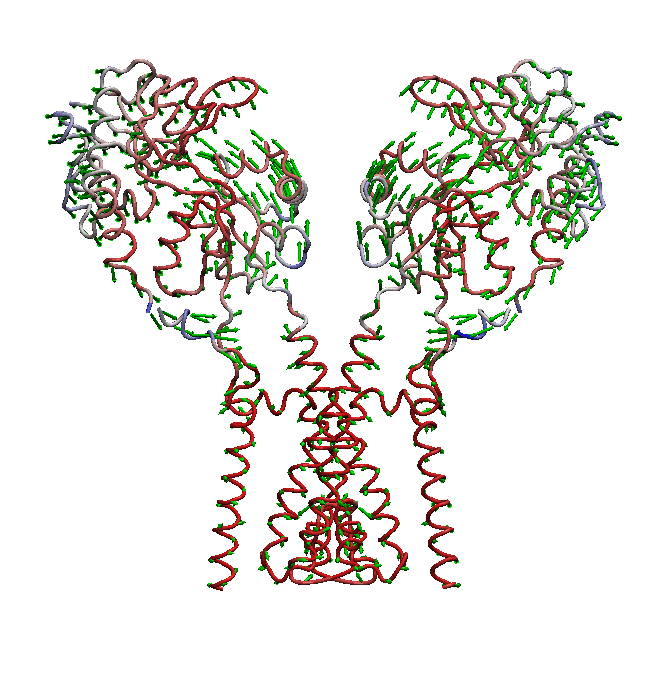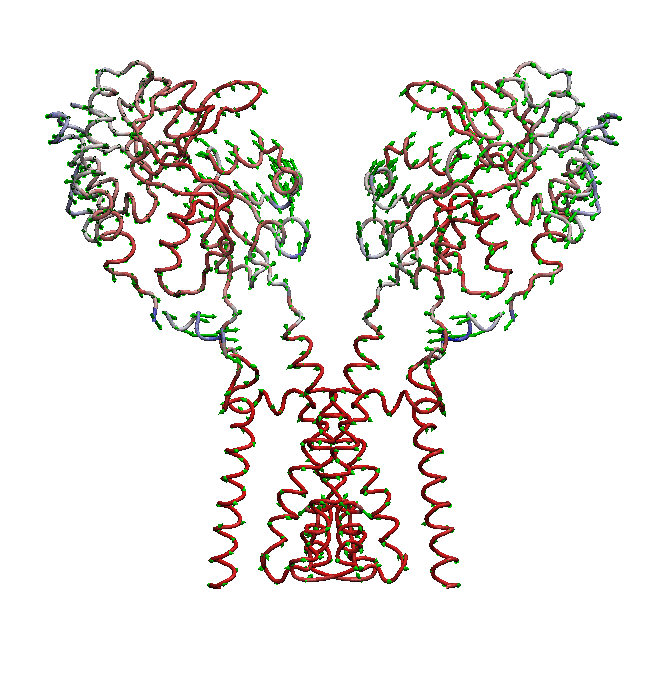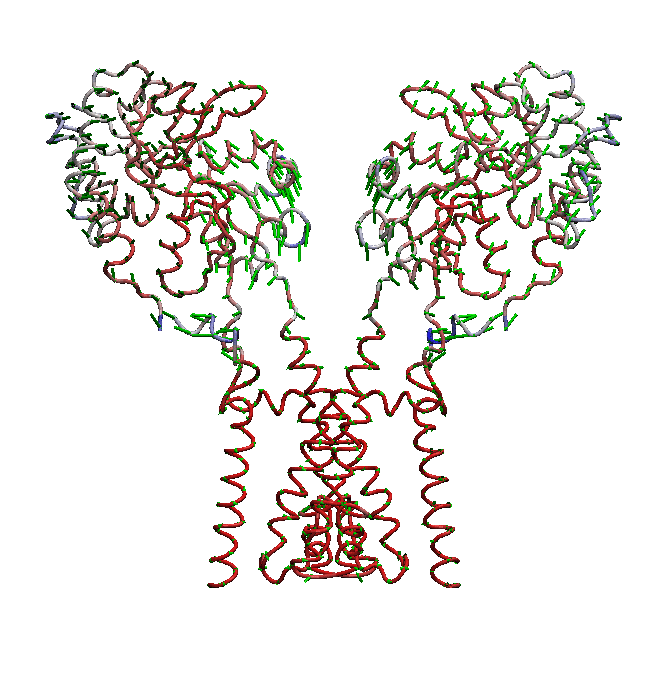
AMPA receptors (AMPARs) are members of the ionotropic glutamate receptor family that mediate fast excitatory neurotransmission in the
central nervous system and are implicated in numerous neurological diseases. Basic architecture of AMPA receptors includes a tetrameric
transmembrane domain (TMD) and a tetrameric ligand binding domain (LBD). While TMD provides the ion permeation pathway and is
responsible for the passage of substrates across the membrane, the cascade of conformational changes that leads to TMD opening of itspore
starts by the glutamate binding in the LBD.
The molecular mechanisms of how the glutamate binding in the ligand binding domain transmits to structural changes in the transmembrane
domain is not well understood. We use molecular dynamics (MD) simulations along with statistical analysis to study molecular bases
of gating (opening and closing of the receptor) in AMPARs. Specifically, our studies help to understand the correlated changes
and structural elements involved in the conformational coupling between the transmembrane domain and the ligand binding domain as the
receptor traverse from apo to open and to desensitized state.
Recent Publications:
M. V. Yelshanskaya, S. Mesbahi-Vasey, M. G. Kurnikova and A. I. Sobolevsky, “Role of the ion channel extracellular collar in AMPA receptor gating”, Scientific Reports 7: 1050 (2017)
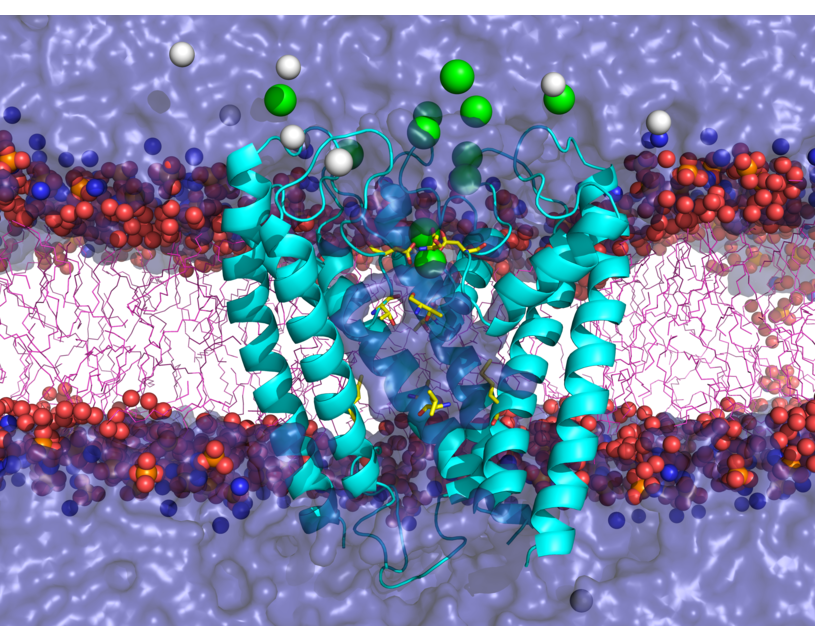
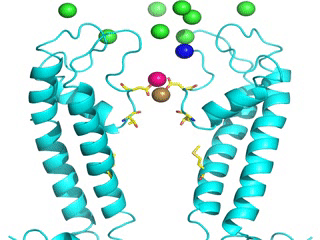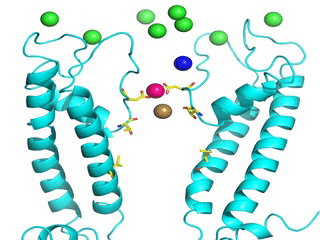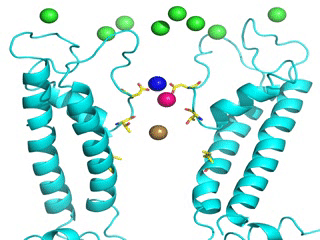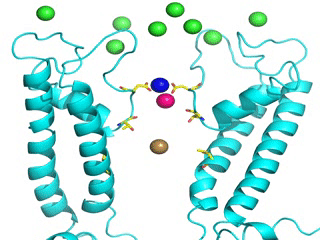
Transient receptor potential (TRP) channels function as sensors for various physical and chemical stimuli and are vitally important players in regulation of key biochemical processes in a variety of cells. TRPV5 and TRPV6 subtypes of TRP channels are the main Ca2+ entry channels in epithelia Their pores are selective for Ca2+ and permeable for a variety of monovalent and divalent cations. An exact mechanism of Ca2+ permeation and selectivity through the TRPV5/6 ion channels is unknown. Recently, a crystal structure of the TRPV6 channel, the first high-resolution structure of a TRP channel, has been solved in complex with different ions bound in the channel pore [1]. Analysis of the structure suggested that Ca2+ permeates TRPV6 channel by a knock-off mechanism. We have created a fully atomistic model of the trans-membrane (TM) domain of the TRPV6 channel in lipid and water, and performed molecular dynamics simulations of Na+ , Ca2+, Ba2+ , and Gd3+ ion permeation through the selectivity filter of this channel. As a result, our MD simulations directly demonstrate the key features of ion permeation through TRPV6 channel pore. In particular, at low calcium concentrations, we observed interplay of Na+ and Ca2+ permeation that follows the knock-off mechanism. We also compare Ca2+ permeation with permeation of Ba 2+ and channel block by Gd3+.
Publications:
Sakipov S, Sobolevsky AI, Kurnikova MG. Ion Permeation Mechanism in Epithelial Calcium Channel TRVP6 Sci Rep. 8(1):5715. (2018)
References:
1. Saotome, Kei, Singh, A. K., Yelshanskaya, M. V., Sobolevsky, A. I. (2016) Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506-511