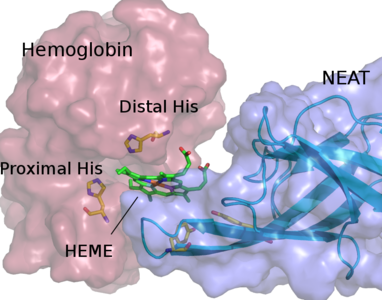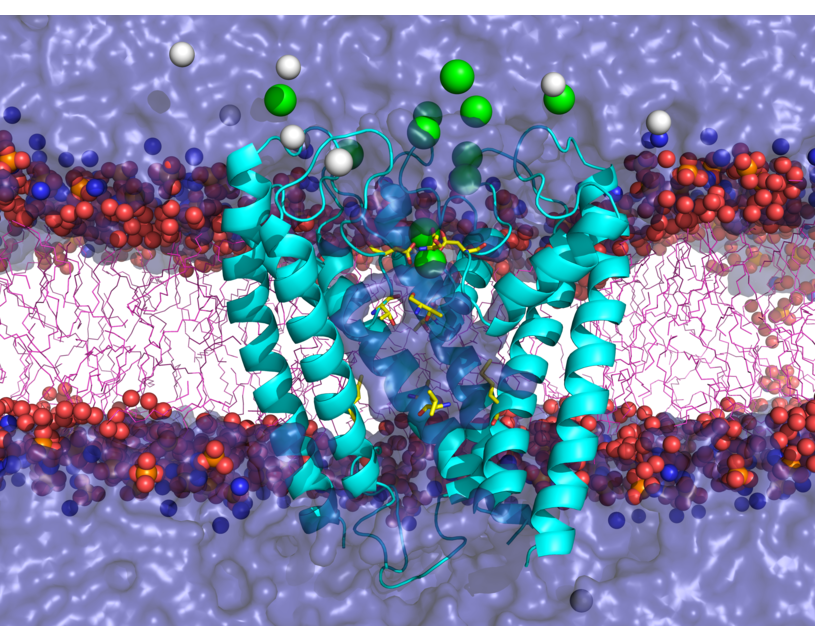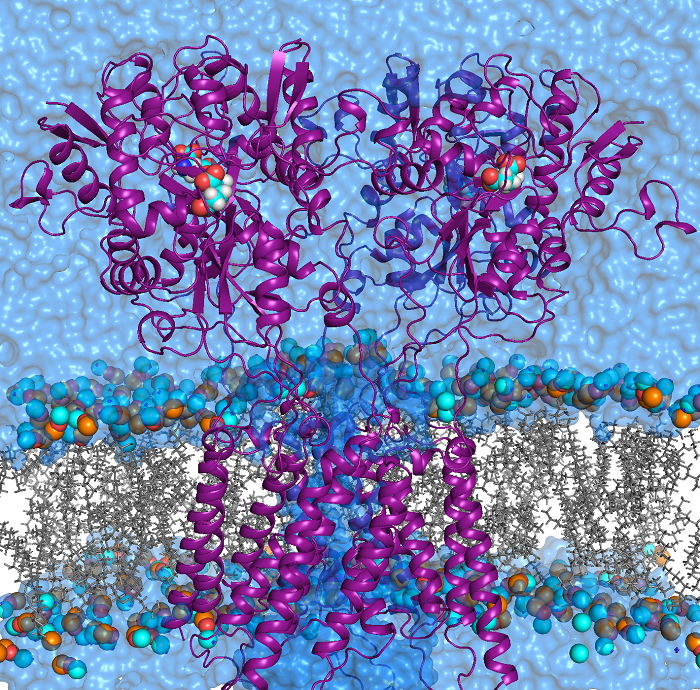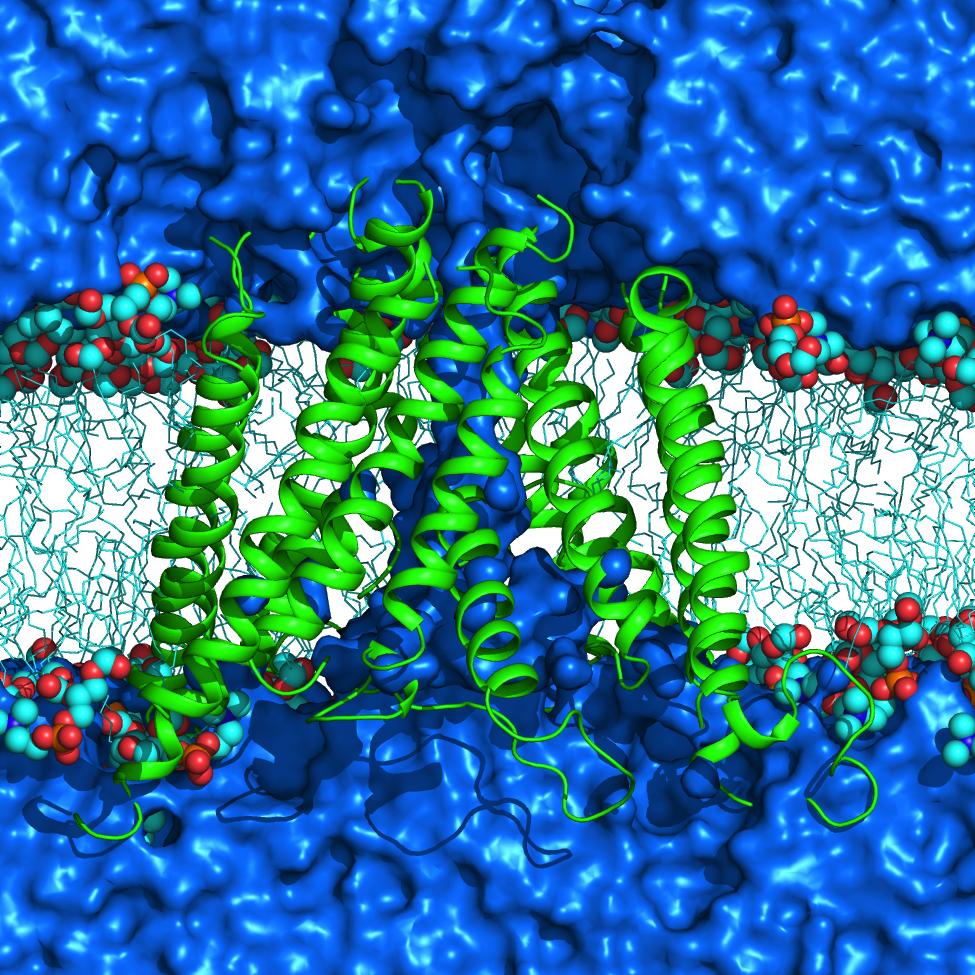
HARLEM (HAmiltonians for Research of LargE Molecules)
HARLEM is a modular molecular modeling and analysis program with .
with a cross-platform GUI. It is interfaced to major molecular modeling engines, such as AMBER, GROMACS and Gaussian.
HARLEM recognizes a range of molecular formats: PDB, XYZ, MOL2, HIN
Python is used for scripting and extensibility.
HARLEM is available for UNIX and WINDOWS operating systems.
The program is is distributed under the open software license
Source code of HARLEM is hosted at GITLAB
Download and Install HARLEM for Windows and above
-
A short manual for HARLEM front end is found here
-
Doxygen documentation found here
Conferences

Department of Chemistry, Carnegie Mellon University https://www.chem.cmu.edu
Computational Biology Department, Carnegie Mellon University http://www.cbd.cmu.edu
Joint CMU-Pitt Ph.D. Program in Computational Biology http://www.compbio.cmu.edu/
Joint CMU-Pitt Ph.D. Program in Molecular Biophysics and Structural Biology http://www.mbsb.pitt.edu
Computational and Systems Biology Department, University of Pittsburgh School of Medicine https://www.csb.pitt.edu/