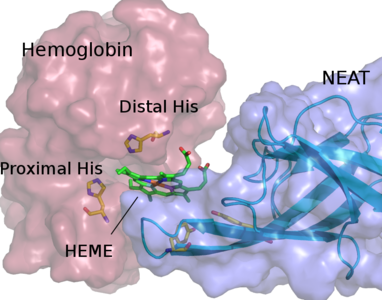
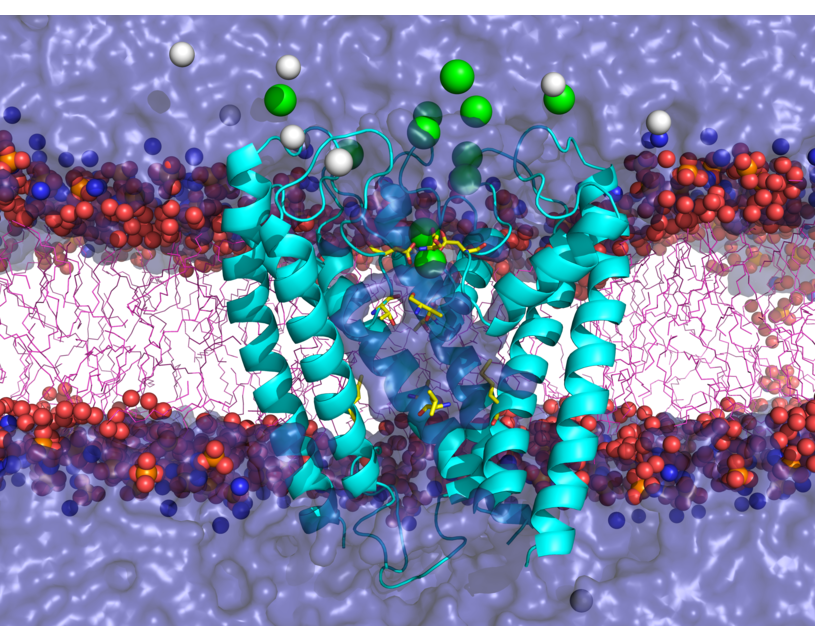
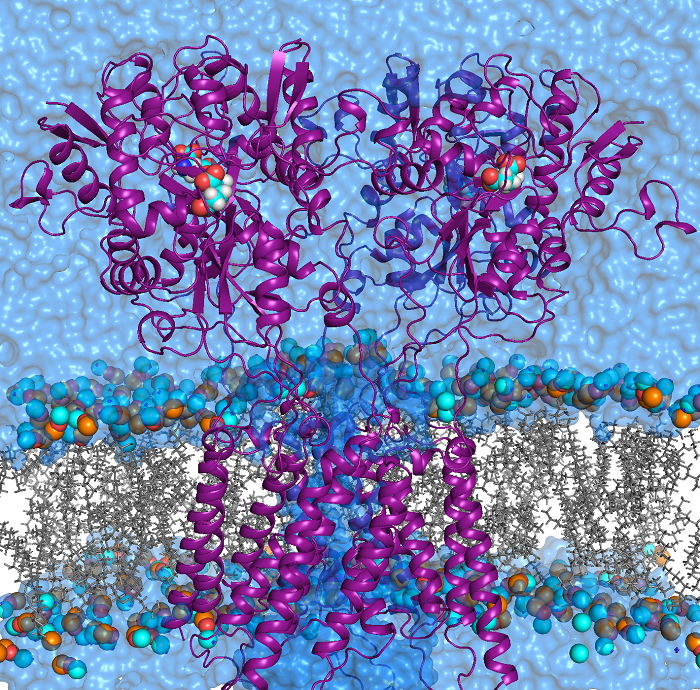
Software developed and maintained by our group:
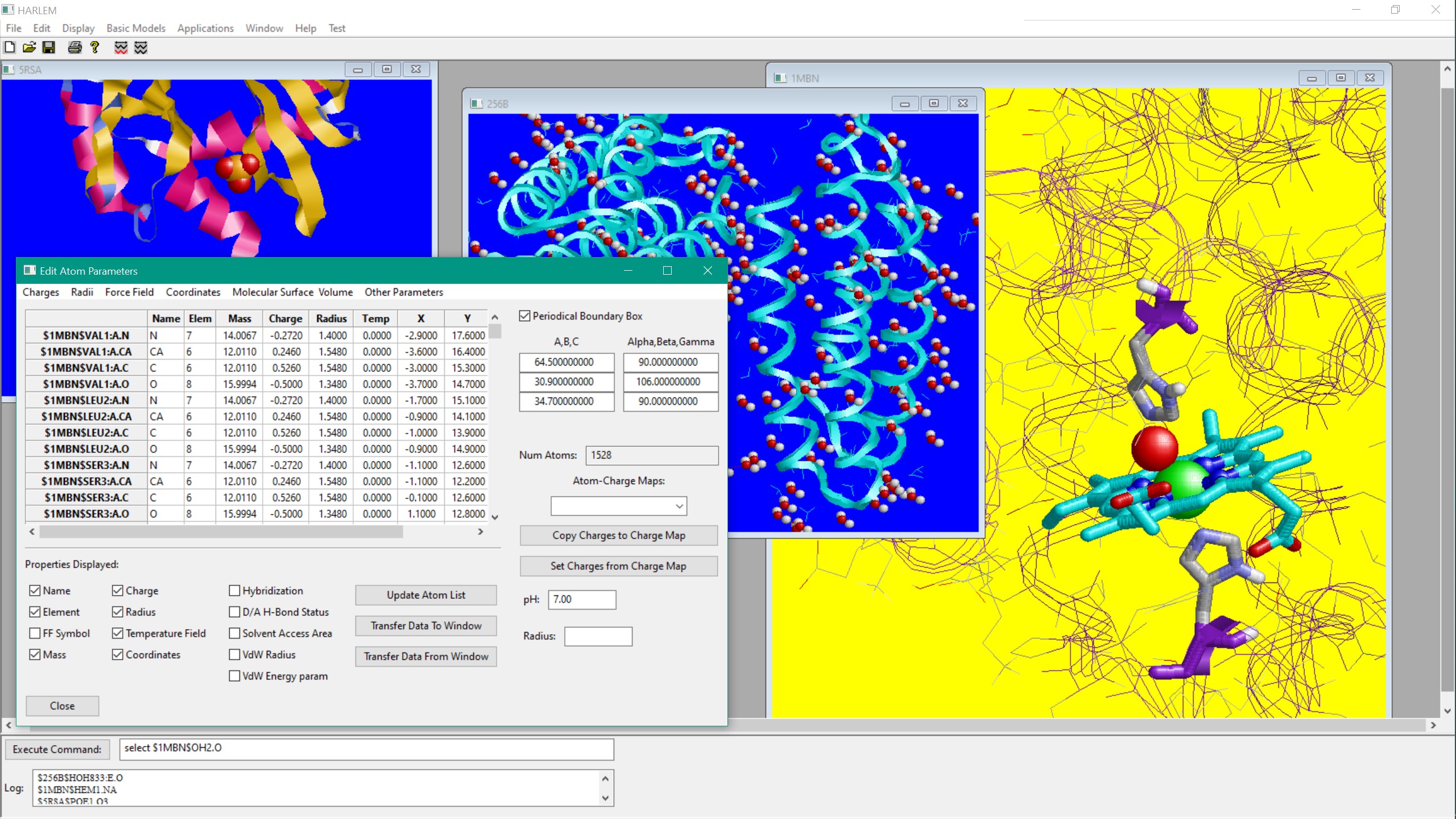
HARLEM - HAmiltonians for Research of LargE Molecules
Harlem is a highly modular multy-representation molecular modeling and analysis tool
with graphical interface (GUI), as well as a front and back ends for the major molecular modeling engines, such as AMBER and Gaussian
HARLEM recognizes a range of molecule formats: PDB, XYZ, MOL2, HIN
Interfaced with Python 3.9 for scripting and extensibility
Software used by our group:
We use a variety of Molecular modeling,visualization, simulation and design software packages:
- AMBER
- Gromacs
- GAUSSIAN 09
- Autodock VINA
- OpenEye
- AMPAC GUI = Gauss View
- Pymol
- VMD
- TensorFlow