Software developed and maintained by our group:
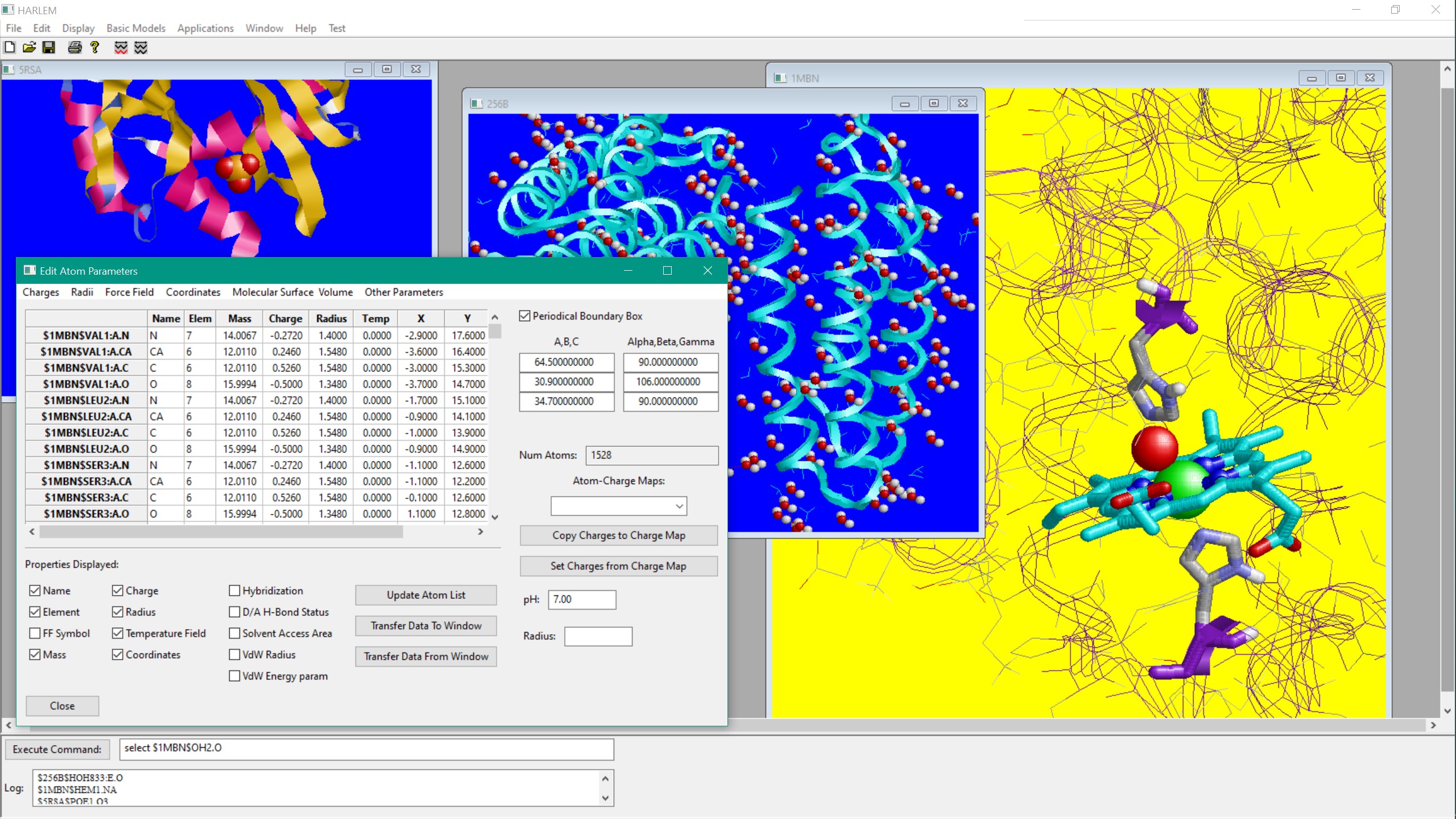
HARLEM - HAmiltonians for Research of LargE Molecules
Harlem is a highly modular multy-representation molecular modeling and analysis tool
with graphical interface (GUI), as well as a front and back ends for the major molecular modeling engines, such as AMBER and Gaussian
HARLEM recognizes a range of molecule formats: PDB, XYZ, MOL2, HIN
Interfaced with Python 3.9 for scripting and extensibility
Software used by our group:
We use a variety of Molecular modeling,visualization, simulation and design software packages:
- AMBER
- Gromacs
- GAUSSIAN 09
- Autodock VINA
- OpenEye
- AMPAC GUI = Gauss View
- Pymol
- VMD
- TensorFlow
Subcategories
List of projects
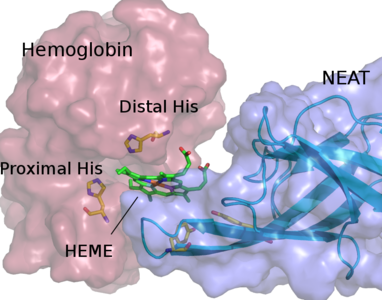
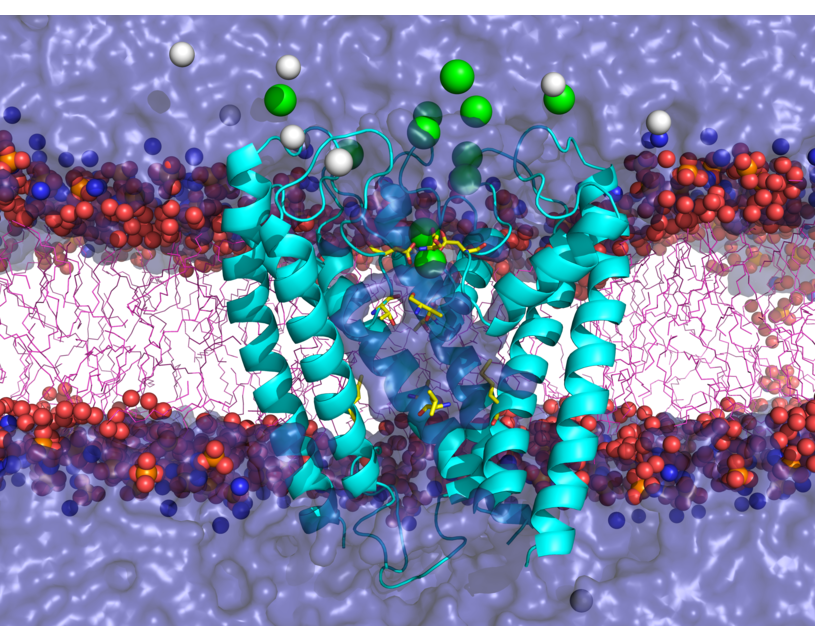
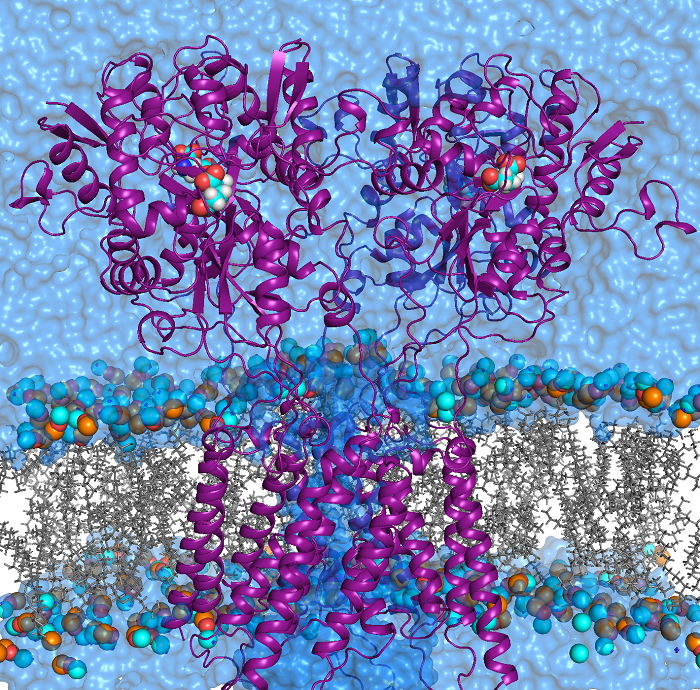
Our group focuses on theoretical/computational modeling of structure/function relationships in biological macromolecules: proteins, lipids and RNA. We study ion selectivity and allosteric mechanisms of gating and modulation in ion channel and receptor proteins. These proteins are typically constructed as multi-subunit assemblies. Each subunit may in turn consist of several structurally and functionally distinct domains assembled in a modular fashion. For example, ionotropic glutamate receptors (iGluRs) are formed as tetramers in which a bundle of alpha-helixes spans lipid bilayer (biological membrane) to form a water-filled channel for ion permeation. The channel is gated (opens and closes) when ligand molecules, such as an agonist glutamate, bind to the extra-cellular ligand binding domains of the protein. The time scales on which such channel may function are of the order of milliseconds. Transmission of a gating signal from the ligand binding site to the transmembrane domain is not well understood. We are working toward developing theoretical models and methodologies which combine molecular level description of the protein and its important “chemistry” (e.g. ligand interaction with the ligand binding domain) and a coarse grained description of interactions and mobility of protein domains and subunits on a micro- to millisecond time-scales relevant to the physiological function.
Below you can find a list of our most recent projects: