 |
Welcome to Kurnikova Research Group
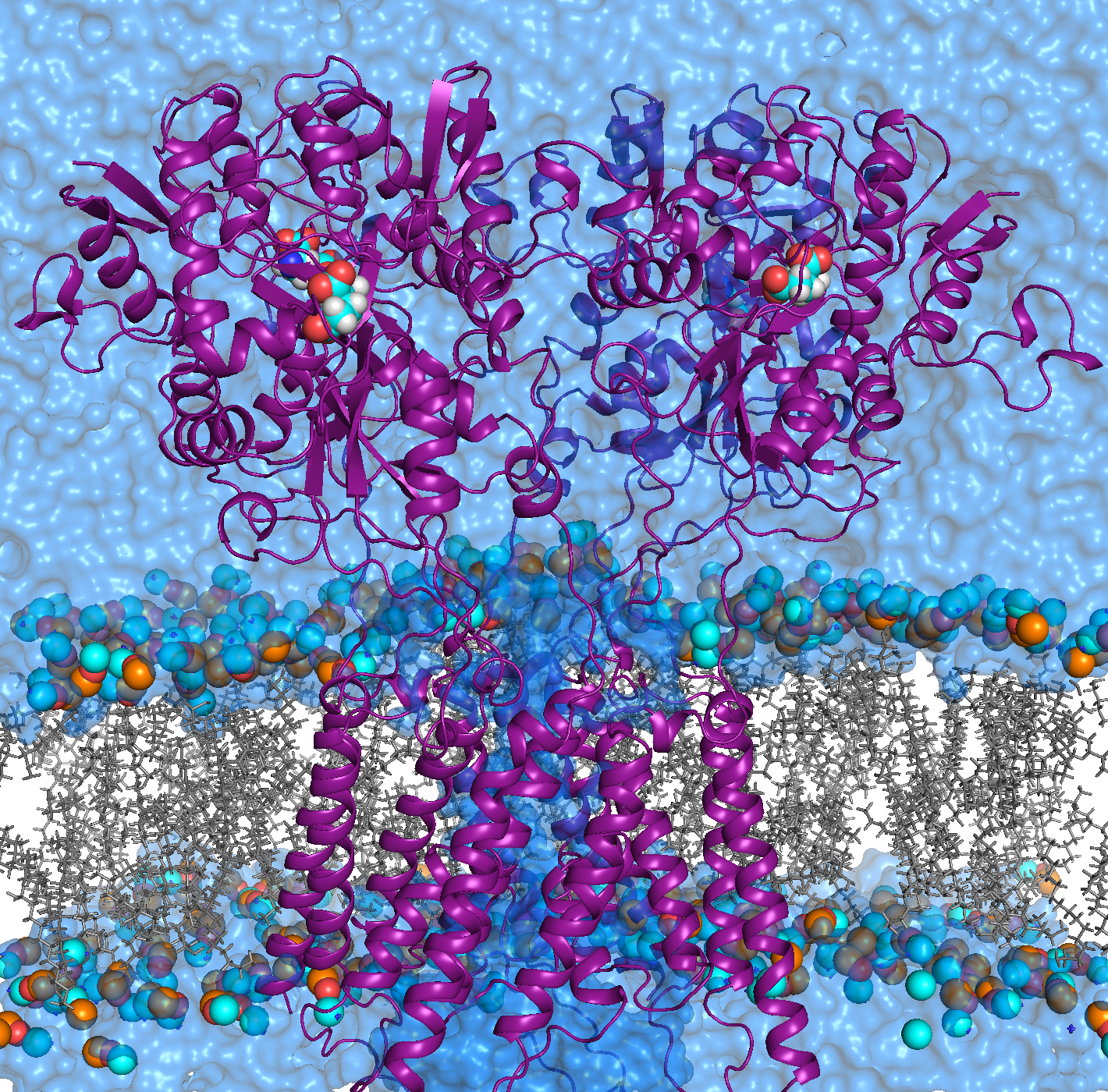
Our group specializes in theoretical computational modeling of structure-function relationships in biological macromolecules, such as proteins, lipids and RNA. |
 |
Welcome to Kurnikova Research Group
Our group specializes in theoretical computational modeling of structure-function relationships in biological macromolecules, such as proteins, lipids and RNA. |
|